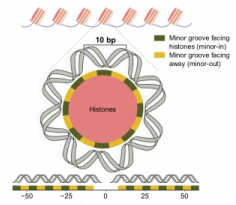
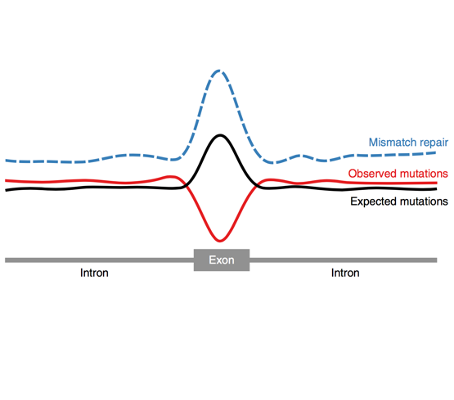
Our paper on how nucleosomes influence mutation rate leading to a 10bp periodic pattern is out in Cell
We are very excited to share that our work describing the somatic and germline 10bp periodicity in the mutation rate within nucleosome-bound DNA was published in the [...]