News
From our Lab
July 30, 2021
Published in Nature our new machine learning method to analyze cancer driver mutations titled “In Silico saturation mutagenesis of cancer genes”.
BoostDM is a computational tool based on machine learning designed to identify driver mutations affecting cancer genes. The method, inspired in evolutionary biology, is designed to define the [...]
December 17, 2020
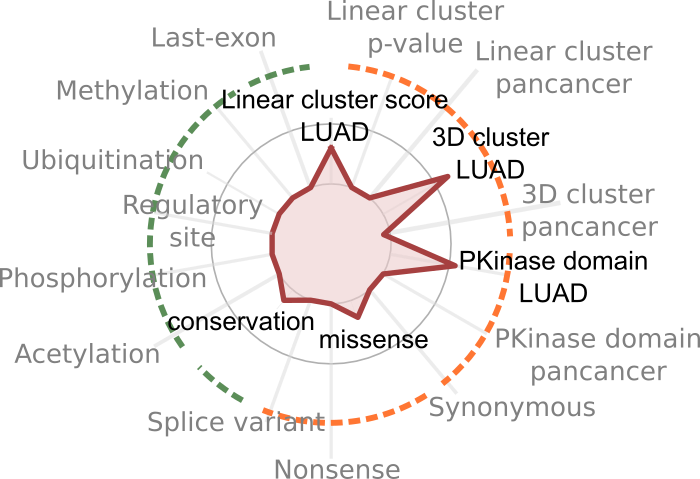
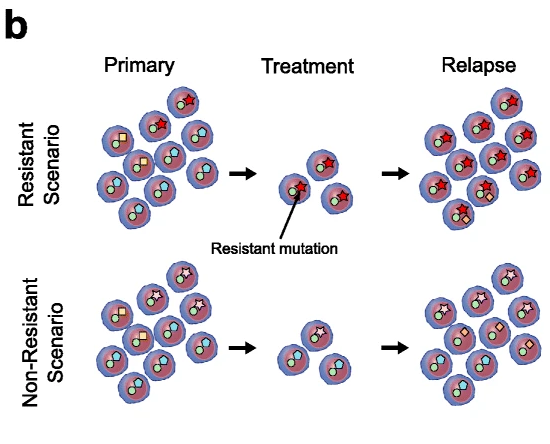
Our study about relapse of T-cell acute lymphoblastic leukemia (T-ALL) in adult patients published in Genome Biology
This work performed in collaboration with Hospital del Mar Medical Research Institute and Josep Carreras Leukaemia Research Institute, and supported by Asociación Española Contra el Cáncer (AECC), [...]
September 2, 2020
Our review describing IntoGen titled “A compendium of mutational cancer driver genes” published in Nature Reviews Cancer
In this review, we focus on the history of cancer driver genes identification from the beginning of cancer research through the advent of cancer genomics and into [...]
December 16, 2019
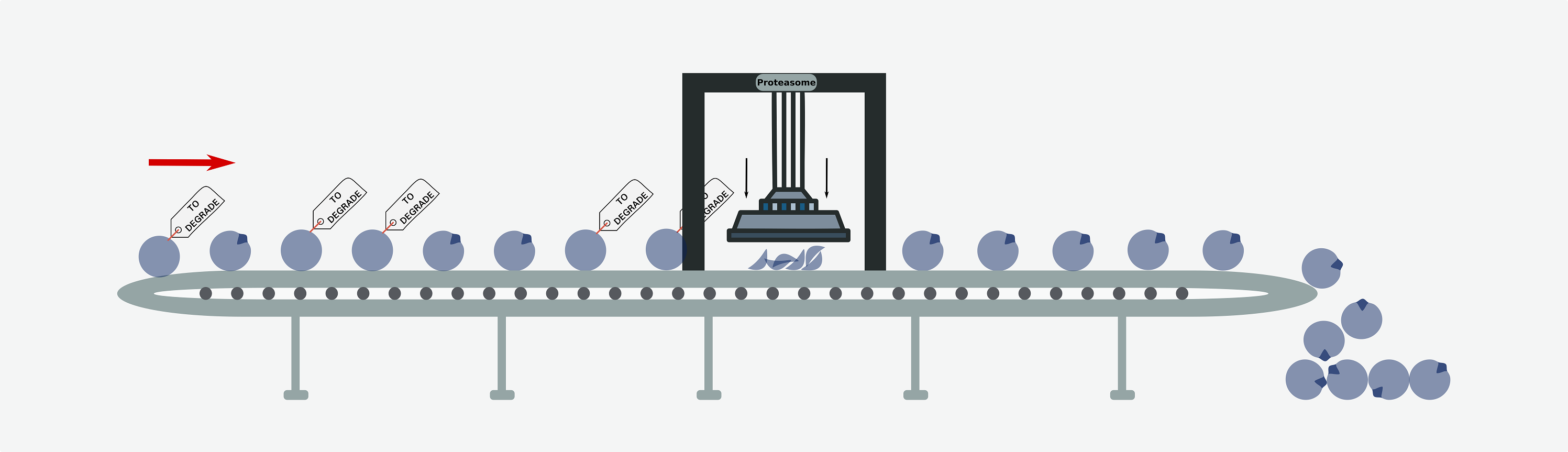
Our work on how alterations of ubiquitin-mediated proteolysis affect in cancer published in Nature Cancer
We are very happy to have published one of the first articles of the Nature Cancer journal. We have systematically analyzed how alterations in the ubiquitin proteolysis system [...]