Our paper demonstrating that proteins bound to DNA impair Nucleotide Excision Repair published in Nature
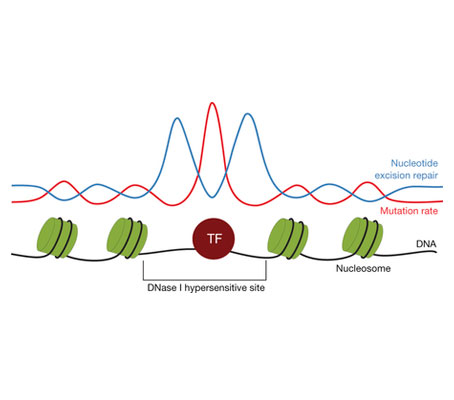
I am very happy to announce that our manuscript describing the increased mutation rate in Transcription Factor Binding Sites (TFBS) in melanomas and lung tumors has been published in today’s issue of Nature. In the manuscript we demonstrate that this accumulation is due to the impairment of Nucleotide Excision Repair (NER) activity by proteins bound to DNA.

Exposure to UV radiation causes specific DNA lesions, which are recognised and repaired by Nucleotide Excision Repair (NER) machinery. Non-repaired lesions may result in mutations during DNA replication. Skin tumors, such as melanomas, carry large number of mutations resulting from these unrepaired DNA lesions. Our study shows that in the DNA regions bound by proteins, such as Transcription Factors (TF), the activity of NER machinery is strongly reduced leading to an increased mutation rate in those sites. By Iris Joval
The manuscript was first published as a preprint in bioRxiv on October 2015
As many researchers have experienced, the editorial and peer-review process may be very long. We submitted the manuscript of this article for the first time to Nature on 9th June 2015, which initially rejected it without peer-review. After various transfers from one journal to another and a first round of peer-review, it went back to Nature in October 2015 for a second round of revision. At that time we felt we couldn’t wait longer to make this finding publicly available to the research community. Many research groups are analyzing tumor whole-genomes with the objective to find non-coding driver mutations. For instance, we are working together with a group of international researchers as part of the ICGC-TCGA PanCancer Analysis of Whole Genomes Project to identify driver mutations in 2800 tumor whole-genomes. The increased mutation rate in TFBS due to faulty repair may easily be mistaken by driver mutation hotspots in promoters, and thus we felt that we had the responsibility to describe our finding as soon as possible to the community. At this point, we felt we couldn’t wait for the long review process, and we decided to publish the manuscript in bioRxiv on October 13th 2015. Since then it has had more than 1000 PDF downloads.
We are grateful to the team maintaining bioRxiv and also to Nature for considering manuscripts previously posted in these repositories.
Reproducible research
To facilitate the full reproducibility of our results, we have published the data and code to carry out all the analyses and results we report in the article. These are available at http://bg.upf.edu/tfbs, where all datasets used can be downladed, and it links to the git repository with the code https://bitbucket.org/bbglab/tfbsmutrate. A sample of the code to reproduce Figure 1A is shown in the image below.



