In silico prescription of anticancer drugs to cohorts of 28 tumor types reveals novel targeting opportunities
We are pleased to announce the publication of our paper in Cancer Cell describing the landscape of anti-cancer targeted therapeutic opportunities across a cohort of patients of twenty eight of the most prevalent cancers.
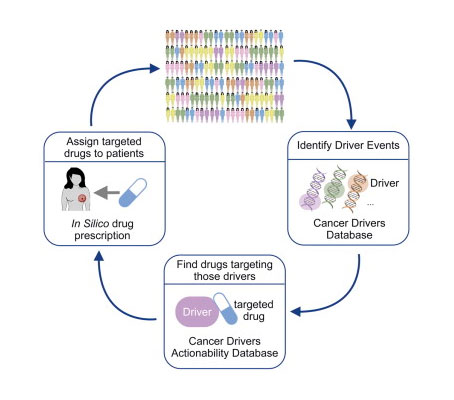
Targeted therapies exploit tumor vulnerabilities offered by specific cancer alterations. However, not all genomic alterations are equally relevant for a given tumor individual. Tumors are more dependent on alterations directly involved in their development and maintenance, namely driver alterations. These represent good options to develop targeted therapies. In this paper we describe an in silico prescription approach to obtain the landscape of targeted drugs against cancer driver alterations, composed of three main steps.
1) Discover the actionable driver events —including somatic mutations, copy number alterations and gene fusions– of each tumor individual. Three distinct signals of positive selection in the pattern of somatic mutations across tumors were traced to detect mutational driver genes in each cancer type. Copy number and fusion drivers acting in the cohort were manually added to this list. Thereafter, we classified these cancer driver genes into Activating and Loss-of-function. All this information was stored in the Drivers Database available at http://www.intogen.org.
2) Gather all available information on therapeutic agents —including FDA approved drugs, and agents in clinical or pre-clinical stages– that target those cancer driver events. We considered direct targeting, indirect targeting and gene therapy strategies. Moreover, we encoded a set of ancillary rules that these agents must fulfil to target the aforementioned driver alterations with different levels of evidence. All this information composed the Drivers Actionability Database, which is also available through IntOGen (http://www.intogen.org/downloads).
3) Finally, by combining data of both the Drivers Database and the Drivers Actionability Database with the alteration data of each tumor individual, we selected -or in silico prescribed- the anti-cancer targeted drugs that could potentially benefit each patient.
In summary, in the Driver Database we identified 475 cancer driver genes acting in one or more tumor types (459 of which are mutational cancer driver genes). Fifty-one of them could be targeted by FDA approved agents and 26 other by molecules currently in clinical trials. We also identified 80 therapeutically unexploited targetable cancer genes, bound by pre-clinical molecules or suitable for small molecule/antibody binding. Lastly, by applying in silico drug prescription we found that only 5.9% of the patients could be treated following current clinical guidelines. However, considering several repurposing strategies that we have proposed, the fraction of patients that could potentially benefit from FDA approved drugs increases up to 40.2%. Moreover, this fraction increases to 73% if we also take into account the patients that could benefit of targeted agents that are currently in clinical trials.
We hope our work enriches the toolbox to interpret tumor genomes, thus supporting the advance of personalized cancer medicine, both by identifying actionable items for targeted drugs and by proposing potential new therapeutic opportunities to solve unmet needs. We are also glad to make available the resources generated as result of this study, i.e. the Cancer Drivers Database and the Drivers Actionability Database, which we hope will be valuable for the scientific community (http://www.intogen.org/downloads/).
Citation:
Carlota Rubio-Pérez, David Tamborero, Michael P. Schroeder, Albert A. Antolín, Jordi Deu-Pons, Christian Perez-Llamas, Jordi Mestres, Abel González-Pérez, Nuria López-Bigas (2015). In Silico Prescription of Anticancer Drugs to Cohorts of 28 Tumor Types Reveals Targeting Opportunities”, Cancer Cell 27, 1–15 March 9, http://authors.elsevier.com/a/



